MAKE A MEME
View Large Image
| View Original: | TMRCAs-compared.PNG (566x349) | |||
| Download: | Original | Medium | Small | Thumb |
| Courtesy of: | commons.wikimedia.org | More Like This | ||
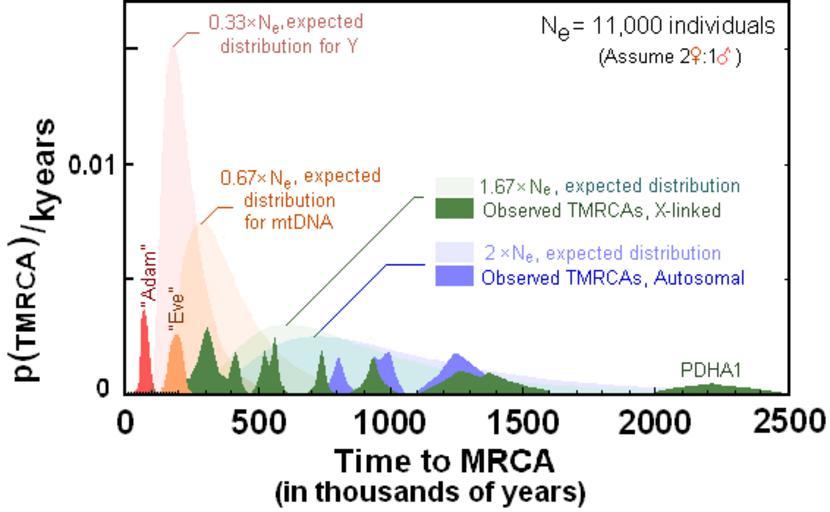
| Keywords: TMRCAs-compared.PNG graph en This figure is largely adopted from Schaffner SF The X chromosome in population genetics Nature Reviews -Genetics 2004 43-51 From figure box 3 panels b-top and bottom Other data added on X-linked loci TMRCAs all adjusted to fit 6MYA C/H LCA This figure differs from Shaffer in the following ways 1 haploid peak broken into two parts as per Wilder JA Mobasher Z Hammer MF Genetic evidence for unequal effective population sizes of human females and males Mol Biol Evol 2004 Nov;21 11 2047-57 male more recent and female less recent 2 For X-linked was 1 666 x Ne as per the above reference instead of 1 5 3 Shaffner used an 12 000 Ne population size were as I use the size suggested by Tanahata however each probability distributions are offset to the right by 75 000 years to deal with the effects of population expansion 4 The TMRCA for mtDNA is added as per Gonder et al 2007 194 000 years +/- 32 000 years 5 PDHA1 gene is added and two recently fixed X-linked genes are added at the recent edge of the probability distribution for ascetic reasons the sites used in these two early studies were two few to establish good statistics and thus the TMRCA would be broad HLA DRB1 locus and MX1 locus are not shown The curves were generated computationally assuming that estimated fixation time as the Sum g p g where g is the the number of generations past and the generation time is 22 years as per gonder Therefore parameters are set to acheive E 2 Ne and generation data following 2N rule was output on excel excluded and fixed own Pdeitiker 09-05-2009 graph Most recent common ancestor Mitochondrial eve Y-chromosomal Adam cc-zero | ||||