MAKE A MEME
View Large Image
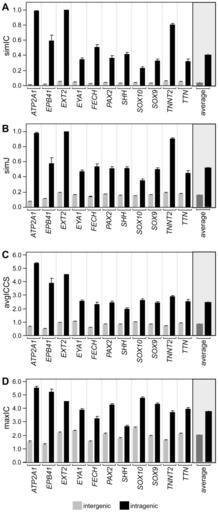
| View Original: | Linking-Human-Diseases-to-Animal-Models-Using-Ontology-Based-Phenotype-Annotation-pbio.1000247.g005.jpg (469x1099) | |||
| Download: | Original | Medium | Small | Thumb |
| Courtesy of: | commons.wikimedia.org | More Like This | ||
| Keywords: Linking-Human-Diseases-to-Animal-Models-Using-Ontology-Based-Phenotype-Annotation-pbio.1000247.g005.jpg en Each of the four panels shows one of the four similarity measurements comparing the score for alleles of the same gene intra in black versus alleles of all other genes inter in gray for each of the 11 OMIM genes annotated The average of all 11 OMIM gene comparisons for each similarity metric are shown in the grayed portion of the graph on the right Metrics are as described in Figure 4 A simIC B simJ C ICCS and D maxIC For each metric there was a significantly higher similarity value p<0 0001 for the intra-genic comparisons as compared to the inter-genic comparisons Significance was tested using a two-tailed Student's t-test for the pairwise comparison intra versus inter for all four metrics for each gene Error bars are standard error of the mean 2009-11 http //dx doi org/10 1371/journal pbio 1000247 Image file from Cite journal Washington N Haendel M Mungall C Ashburner M Westerfield M Lewis S Linking Human Diseases to Animal Models Using Ontology-Based Phenotype Annotation 10 1371/journal pbio 1000247 PLOS Biology 2009 19956802 2774506 Washington N Haendel M Mungall C Ashburner M Westerfield M Lewis S cc-zero Information field Provenance Recitation-bot Synthetic biology Disease models Functional genomics Gene discovery Media from PLOS Biology Uploaded with reCitation Bot Uploaded_with reCitation Bot and needing category review | ||||